(Lead: Alexander Marson; Co-I: Jennifer Doudna, Harmit Malik, Michael Emerman, Judd Hultquist & Nevan Krogan)
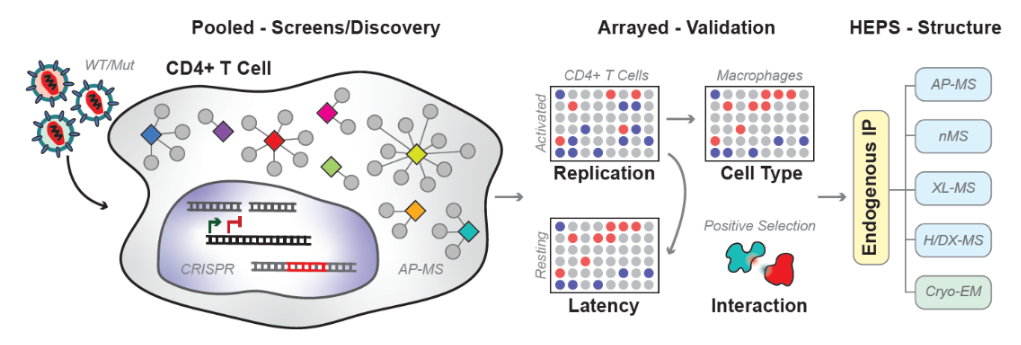
Host-virus interfaces of well-described effector complexes have been structurally characterized based on lab-adapted strains of HIV and single host sequences found in cell line models of HIV infection. However, cell lines do not reflect the functionally distinct molecular biology of HIV infections in primary CD4 T cells and macrophages. Moreover, HIV is one of the most diverse human pathogens, yet most of the available structural data are based on a handful of lab-adapted strains and not primary isolates, and host-pathogen protein interfaces are known to change in evolutionary history. In Project 3, we will investigate host and virus diversity by using primary cells isolated from multiple donors and infecting them with diverse HIV strains, to then identify novel strain-specific proviral and antiviral host factors from a variety of genome-wide pooled screens. In collaboration with the Genetics Core, we will apply a suite of innovative technologies in gene editing of human primary cells, to investigate restriction factors that are under positive selection and identify interactions important for HIV restriction.
Our project will identify the universe of functionally relevant HIV effectors using genome-wide, pooled CRISPRko, CRISPRi and CRISPRa screening in primary CD4 T cells. These results will be integrated with previous datasets to define functional HIV-host networks (with the Computational Core). Novel host factor candidates will be validated by multiplexed, arrayed editing in primary activated CD4 T cells, generating high-confidence interactors that will be further investigated by arrayed editing in resting CD4 T cells to examine their role in HIV latency. To define the function and structure of host complexes required in primary CD4 T cells and macrophages, we will perform high-throughput arrayed CRISPR-Cas9 gene editing of functionally validated factors and infect both cell types with different viral strains and mutants. Protein interaction networks of specific effector proteins will be further analyzed through the HARC endogenous protein structure (HEPS) platform (in collaboration with the Proteomics and Structural Biology Cores). Furthermore, we will map evolutionary and structural constraints by mutational scanning of host-pathogen protein interfaces known to be changing over time. Residues under positive selection will be assessed for position on protein surfaces and known complex interfaces and subjected to mutational scanning in primary T cells by knock-in of polyclonal repair libraries (Genetics Core). In summary, Project 3 will identify novel HIV-host factor complexes for structural interrogation that can be therapeutically targeted and reveal new insights in viral evolution, transmission, and pathogenesis.
The HARC Center projects are supported by 6 technology cores, which are essential for their completion: (1) Proteomic Approaches to HIV Function, (2) CRISPR in Primary Cells to Study HIV-Host Function, (3) Structural biology using Cryo-Electron Microscopy and X-ray Screening, (4) Integrative Modeling of HIV-Human Complexes, (5) Administrative core and (6) development core.