(Lead: Alan Frankel; Co-I: Natalia Jura, Lillian Cohn, Yifan Cheng, Nancie Archin, Geeta Narlikar & Nevan Krogan)
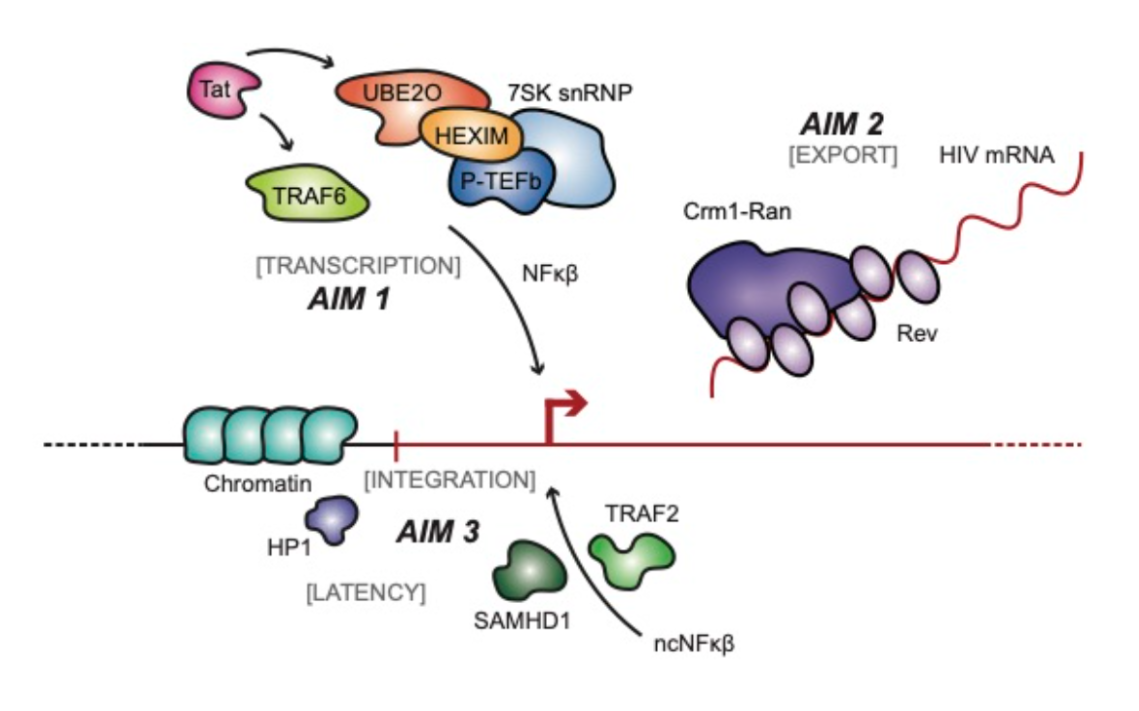
The HIV regulatory proteins Tat and Rev play a crucial role in controlling HIV transcription and mRNA export, respectively. In Project 2, we will functionally and structurally elucidate the roles of ubiquitin ligases that modify Tat, reveal the architecture of the Rev/RRE nuclear export complex to understand the mechanisms regulating HIV mRNA translation, and compare Rev and RRE evolution in SIV and HIV to identify viral factors that contribute to zoonosis. To achieve this, we will determine the functional interactions between the E3 ligases UBE2O and TRAF6 with Tat by cryo-EM or X-ray crystallography (Structural Biology Core) and investigate the mechanism by which UBE2O promotes the release of the inhibitory 7SK snRNP by ubiquitination of the HEXIM1 subunit (Aim 1). We will determine the high-resolution structure of the Rev/RRE/Crm1/RanGTP nuclear export complex by cryo-EM and define genetic and protein interaction landscapes of the Rev/RRE complex (Aim 2). We will also compare its protein-protein and protein-RNA interfaces between HIV-1, HIV-2, and SIV complexes. We will use the HEPS platform (Genetics core, Proteomics core and Structural biology core) and deep mutational scanning to functionally validate these interactions and define them biochemically and structurally.
The latent HIV reservoir represents a significant roadblock to eradicating infection. We aim to uncover factors that drive HIV latency, including virus integration sites and the states of chromatin and chromatin interacting proteins (Aim 3). Using CRISPR-Cas9 (Genetics core), we will integrate a minimal HIV-1 LTR reporter into CD4 T cells to address how integration into specific genomic loci may permit expansion of particular clonal cells without virus expression, and understand mechanistically how a large fraction of latent cells expand in chronically infected individuals over time. To elucidate how the state of chromatin and heterochromatin impact HIV transcription, we will also identify post-translational modifications on HP1 proteins, test the roles of specific HP1 modifications on their phase-separation properties, and assess site-specific HP1 mutants for virus replication. Finally we will examine the role of non-canonical (nc) NF-kB in enhancing latency reversal for shock-and-kill therapies, and test synergies with other latency reversal agents in monocytic and lymphocytic latent cell models, resting CD4 T cells (the major reservoir of latent HIV), and gut macrophages isolated from people living with HIV (PLWH). In summary, Project 2 will uncover potential new interfaces as HIV drug targets and evaluate latency reversal or induction of deep latency for cure strategies.