(Lead: John Gross; Co-I: Michael Emerman, Nevan Krogan, Yifan Cheng & Charles Craik)
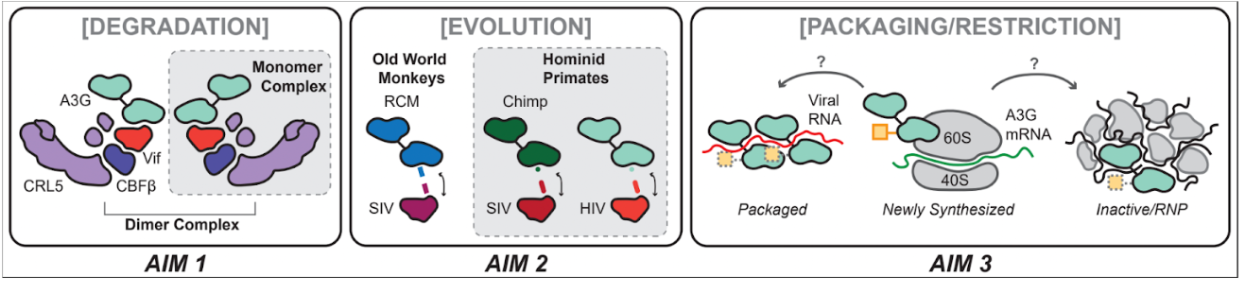
In Project 1 we will elucidate novel structural aspects of the APOBEC3 (A3) family of restriction factors and how they are antagonized by the HIV accessory protein Vif. Primate Vif targets A3’s for degradation by the 26S proteasome, but it is unknown how Vif intercepts A3 packaging complexes. It has been suggested that Vif binds different A3 family members through three different interfaces, but whether these binding sites are independent or dependent on one another is unclear. In previous studies, we uncovered that Vif forms functional interactions with additional host factors, including regulatory subunits of PP2A, components of the chromatin-modifying and transcriptional machinery, and regulators of ubiquitin-mediated proteolysis. We will now investigate how Vif neutralizes different A3 family members to promote efficient viral replication, and how adaptations in Vif enabled neutralization of A3G in hominid primates and how these adaptations affect the ability of human A3G to escape HIV-1.
We will determine the structure of Vif-A3G and use deep mutational scanning to see if there is a tradeoff between binding multiple APOBEC3s (primary cells) and single APOBEC3s (cell lines) or PP2A (P1-Aim 1). We will examine via deep mutational scanning of APOBEC3G positive selection loop its ability to escape HIV Vif (P1-Aim 2). We will use endogenous tagging of host APOBEC3s, identify PPIs and functionally map those that are essential for packaging and solve structures of these functional complexes potentially with RNA bound (P1-Aim 3).
Project 1 will enable rational drug design to target HIV-1 from establishing replication-competent proviruses by utilizing the restriction potential of A3 family members.